Over the years, large-scale language models (LLMs) have emerged as a groundbreaking technology with great potential to revolutionize many aspects of healthcare. These models, such as GPT-3, GPT-4, and MED-PALM 2, demonstrate outstanding capabilities in the understanding and generation of human-like texts, making them valuable tools for tackling complex medical tasks and improving patient care. They show particular promises in a variety of medical applications, including medical questions (QA), dialogue systems, and text generation. Furthermore, with the exponential growth of electronic health records (EHRS), medical literature and patient-generated data, LLM helps healthcare professionals extract valuable insights and make informed decisions.
However, despite the immense possibilities of large-scale language models (LLMs) in healthcare, there are important and specific challenges that need to be addressed.
If the model is used for the conversational aspect of recreation, errors have little effect. This is not the case for use in the medical domain, but incorrect explanations and answers can have serious consequences for patient care and outcomes. The accuracy and reliability of the information provided by the language model can be a matter of life and death as it can affect healthcare decisions, diagnosis, and treatment plans.
For example, if a medical query (see below) was given, GPT-3 incorrectly recommended tetracycline in pregnant patients, but correctly explains contraindications due to potential harm to the fetus. Acting on this false recommendation can lead to bone growth problems in your baby.

To fully leverage the power of LLM in healthcare, it is important to develop and benchmark models using a set up specifically designed for the medical domain. This setup requires consideration of the unique characteristics and requirements of healthcare data and applications. The development of methods for assessing medical LLM is practically important, not just academic interest, given the real risks posed in the medical field.
The Open Medical-LLM leaderboard aims to address these challenges and limitations by providing a standardized platform for assessing and comparing the performance of various major language models across diverse medical tasks and datasets. The Leaderboard aims to promote the development of more effective and reliable medical LLMs by providing a comprehensive assessment of the medical knowledge and question-answer capabilities of each model.
This platform allows researchers and practitioners to identify the advantages and disadvantages of various approaches, promote further advancements in the field, and ultimately contribute to improving patient care and outcomes.
Datasets, Tasks, and Evaluation Setup
The Medical-LLM leaderboard includes a variety of tasks, using accuracy as the primary assessment metric (accuracy measures the percentage of correct answers provided by the linguistic model across different medical QA datasets).
Medqa
The MEDQA dataset consists of multiple choice questions from the US Medical License Examination (USMLE). It covers general medical knowledge, with 11,450 questions in the development set and 1,273 questions in the test set. Each question has four or five answer choices, and the dataset is designed to assess the medical knowledge and reasoning skills required for a US medical license.

Medmcqa
MEDMCQA is a large multi-select QA data set derived from the Indian Medical Entrance Examination (AIIMS/NEET). Covering 2.4k healthcare topics and 21 medical subjects, there are over 187,000 questions in the development set and 6,100 questions in the test set. Each question has four answer choices, accompanied by explanation. MEDMCQA assesses the model’s general medical knowledge and reasoning ability.

PubMedqa
PubMedQA is a closed domain QA dataset that allows you to answer each question by looking at the relevant context (PubMed Abstract). It consists of 1,000 expert-signed question answer pairs. Each question comes with a PubMed Abstract as a context. The task is to provide yes/no/perhaps answers based on the information in the summary. The dataset is split into 500 questions for development and 500 for testing. PubMedqa evaluates the ability of models to understand and reason the scientific biomedical literature.

MMLU Subset (Medicine and Biology)
The MMLU benchmark (measurement of understanding of large-scale multitasking languages) includes multiple choice questions from various domains. The Open Medical-LLM leaderboard focuses on the subset most relevant to medical knowledge.
Clinical Knowledge: Evaluation of 265 clinical knowledge and decision-making skills. Medical Genetics: 100 questions covering topics related to medical genetics. Anatomy: 135 Questions to assess human anatomy knowledge. Specialist Medicine: 272 Questions Assessment of the knowledge required of a medical professional. University Biology: 144 questions covering university-level biology concepts. University Medicine: 173 questions assessing university-level medical knowledge.
Each MMLU subset consists of multiple choice questions with four answer options and is designed to evaluate an understanding of a particular medical and biological domain model.

The Open Medical-LLM leaderboard provides a robust assessment of the performance of the model across various aspects of medical knowledge and reasoning.
Insights and analysis
The Open Medical-LLM leaderboard evaluates the performance of various large-scale language models (LLMs) for diverse medical question tasks. Here are our important findings:
Commercial models such as GPT-4-based and MED-PALM-2 consistently achieve high-precision scores across a variety of medical data sets and demonstrate strong performance in a variety of medical domains. Open source models such as Starling-LM-7B, Gemma-7B, Mistral-7B-V0.1 and Hermes-2-Pro-Mistral-7B show competitive performance on specific datasets and tasks despite having a small size of approximately 7 billion parameters. Both commercial and open source models work well in tasks such as understanding and reasoning around the scientific biomedical literature (PubMedQA) and apply clinical knowledge and decision-making skills (MMLU clinical knowledge subset).
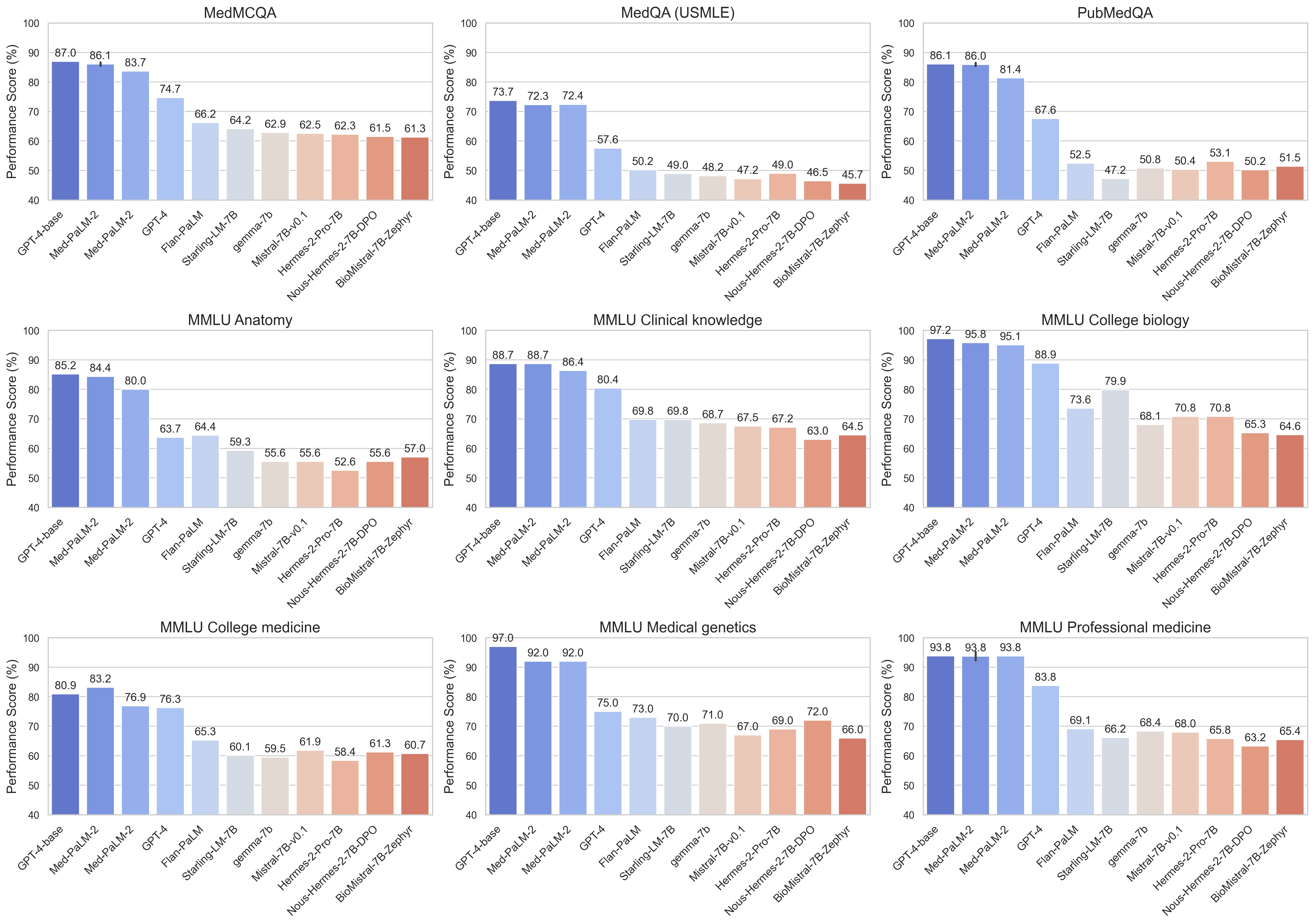
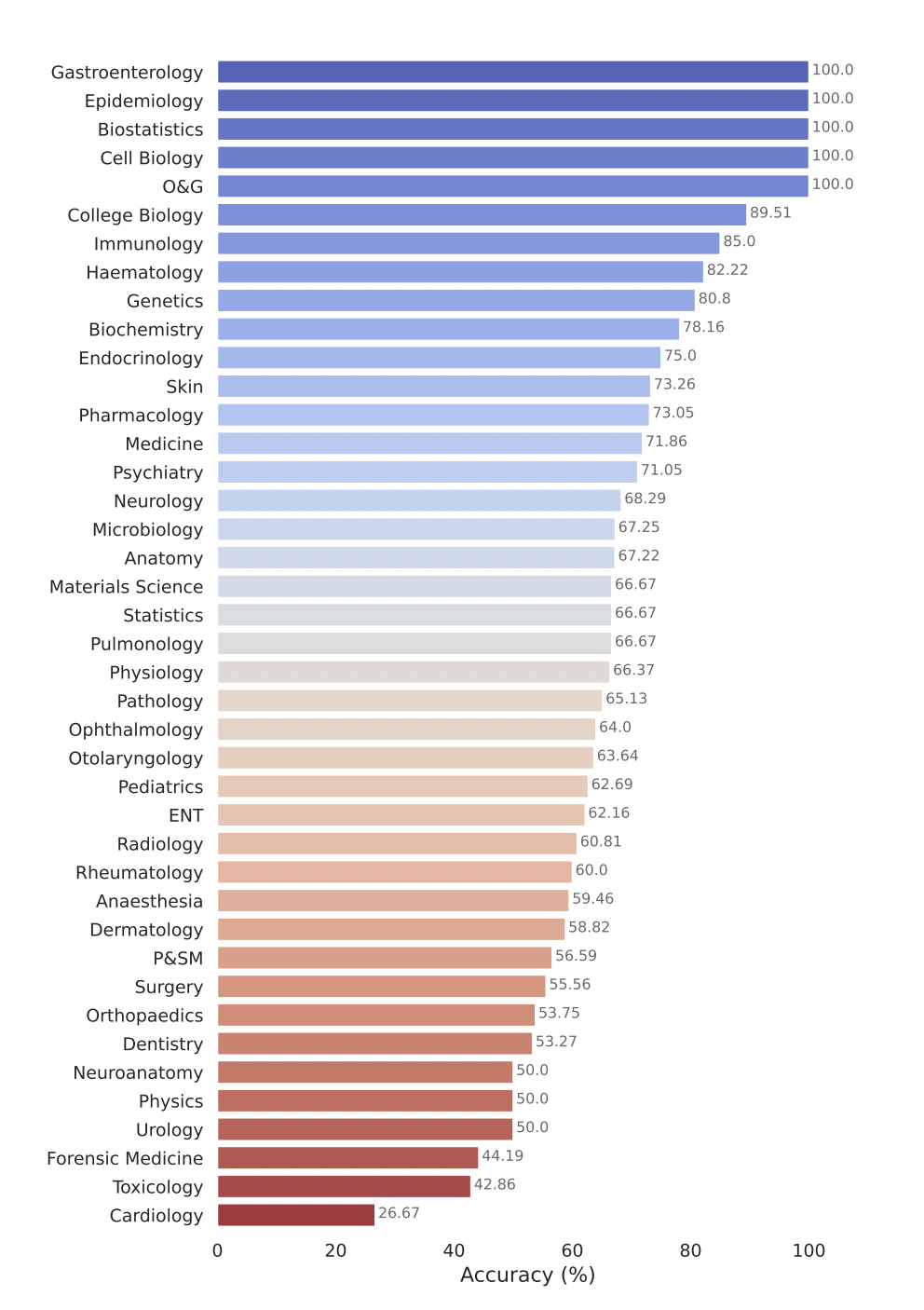
Google’s model, Gemini Pro, demonstrates strong performance across a variety of medical domains. It is particularly excellent at data-intensive and procedural tasks such as biostatistics, cell biology, and obstetrics and gynecology. However, moderate to low performance has been shown in key areas such as anatomy, cardiology, and dermatology, revealing gaps that need to be further refined for comprehensive medical applications.
Submit a model for evaluation
To submit a model for evaluation on the Open Medical RLM leaderboard, follow these steps:
1. Converts model weights to safetenser format
First, convert the weights of the model into Safetensors format. SafeTensors is a new format for storing weights that are safer and faster to load and use. By converting the model to this format, the leaderboard can also display the number of parameters in the model in the main table.
2. Ensure compatibility with Auto Characters
Before submitting a model, make sure you can load the model and talknaser using AutoCharacter from the Trans Library. Use the following code snippet to test compatibility:
from transformer Import autoconfig, autotokenizer config = autoconfig.from_pretrained(model_hub_id) model = automodel.from_pretrained(“Your model name”) tokenizer = autotokenizer.from_pretrained(“Your model name”))
If this step fails, follow the error message to debug the model before submitting it. The model may be uploaded inappropriately.
3. Publish the model
Make sure the model is published. Leaderboards cannot evaluate private models or require special access.
4. Remote code execution (coming soon)
Currently, the Open Medical-LLM leaderboard does not support models that require use_remote_code=true. However, the Leaderboard team is actively working on adding this feature, so stay tuned for the latest updates.
5. Submit the model from the Leaderboard website
If your model is in Safetensors format, compatible with AutoCharacters and is published, you can use “Send Here!” to submit for evaluation. Panel on the Open Medical-LLM Leaderboard website. Fill in the required information, including the model name, description, and additional details, and then click the (Submit) button.
The leaderboard team processes submissions and evaluates the performance of the model on a variety of medical QA data sets. Once the evaluation is complete, the model’s scores will be added to the leaderboard, allowing you to compare your performance with other submitted models.
What’s next? Enlarge the Open Medical LLM Leaderboard
The Open Medical-LLM Leaderboard is committed to expanding and adapting to meet the evolving needs of the research community and the healthcare industry. The key areas of focus are:
Incorporates a wide range of medical datasets covering a variety of aspects, including radiology, pathology, and genomics, through collaboration with researchers, healthcare organizations and industry partners. Enhance evaluation metrics and reporting capabilities by investigating additional performance measures beyond accuracy, such as point-wise scores and domain-specific metrics that capture unique requirements for medical applications. Several efforts are already underway in this direction. If you’re interested in collaborating on the next benchmark you’re planning to propose, join the Discord community to learn more and get involved. We would like to work together to brainstorm ideas!
If you are interested in the intersection of AI and healthcare, building models for the healthcare domain, and the safety and hallucination issues of medical LLM, we recommend joining a disagreeable and vibrant community.
Credits and Acknowledgements

Thank you to all those who made this possible, including the Clémentine Fourrier and the Hugging Face team. We would like to thank Andreas Motzfeldt, Aryo Gema and Logesh Kumar Umapathi for discussion and feedback on the leaderboards under development. We would like to express our sincere gratitude to Professor Pasquale Minervini for his time, technical assistance, and GPU support from the University of Edinburgh.
About Open Life Science AI
Open Life Science AI is a project aimed at revolutionizing the application of artificial intelligence in the life sciences and healthcare domains. It serves as a central hub for lists of healthcare models, datasets, benchmarks, and meeting deadline tracking, and encourages collaboration, innovation and advancement in the field of AI-assisted healthcare. We strive to establish Open Life Science AI as the best destination for those interested in the intersection of AI and healthcare. We provide a platform for researchers, clinicians, policy makers and industry experts to engage in dialogue, share insights and explore the latest developments in the field.

Quote
If you find our ratings helpful, consider quoting our work
Medical-LLM Leaderboard
@misc {Medical-llm Leaderboard, Authors = {Ankit Pal, Pasquale Minervini, Andreas Geert Motzfeldt, Aryo Pradipta Gema and Beatrice Alex}, Tittle = {openlifescienceai/Open_medical_llm_leaderboard}, “\url {https://huggingface.co/spaces/openlifescienceai/open_medical_llm_leaderboard}”}



