![]()

Protein Language Models (PLMs) have emerged as a powerful tool for predicting and designing protein structure and function. At the international conference on Machine Learning 2023 (ICML), MILA and Intel Labs have released Protst, a pioneering multimodal language model for protein design based on text prompts. Since then, Pridst has been well received in the research community, accumulating over 40 citations within a year, indicating the scientific strength of the work.
One of the most popular tasks in PLM is to predict the intracellular position of amino acid sequences. In this task, the user supplies the model with an amino acid sequence, and the model outputs a label indicating the intracellular position of this sequence. Out of the box, the Zero Shot Protst-ESM-1B is better than the state-of-the-art shot classifiers.
To make PRITST more accessible, Intel and Mila reorganized and shared the model with a hugging face hub. You can download the model and dataset from here.
This post shows you how to efficiently perform Prutst inference and fine-tune it with the Intel Gaudi 2 accelerator, as well as the best way to use the Intel Gaudi open source library. The Intel Gaudi 2 is a second-generation AI accelerator designed by Intel. Check out our previous blog post for a detailed introduction and a guide to accessing it via Intel Developer Cloud. Thanks to the best of Intel Gaudi Library, you can port trans-based scripts to Gaudi 2 with minimal code changes.
PRITST’s reasoning
Common intracellular locations include the nucleus, cell membrane, cytoplasm, mitochondria, and more, as described in this dataset.
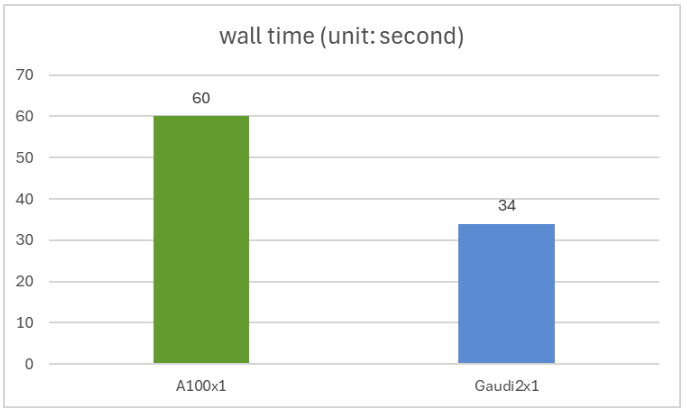
Compare PROTST inference performance on NVIDIA A100 80GB PCIE and Gaudi 2 accelerators using test splitting of Protst-sublularLocalization Dataset. This test set contains a 2772 amino acid sequence with variable sequence lengths ranging from 79 to 1999.
You can use this script to replicate your experiment. This script runs the model with full BFLOAT16 accuracy with batch size 1. The NVIDIAA100 and Intel Gaudi 2 have an accuracy of 0.44, with the Gaudi2 1.76 times faster estimate speed than the A100. The wall times for a single A100 and a single Gaudi 2 are shown in the diagram below.
Finely tuned protrusion
Fine-tuning the protost model in downstream tasks is a simple and established way to improve modeling accuracy. This experiment specializes in models of binary positions in simpler versions of subcellular localization using binary labels indicating whether the protein is membrane bound or soluble.
You can use this script to replicate your experiment. Here we will fine-tune the Protst-ESM1B-For-Sequential-Classification model with BFLOAT16 accuracy in Protst-BinaryLocalization Dataset. The table below shows the model accuracy of test splitting using various training hardware setups, closely in line with the results published in the paper (approximately 92.5% accuracy).

The diagram below shows the fine tuning times. The single Gaudi 2 is 2.92 times faster than the single A100. This diagram shows how the distributed training scale is roughly linearly for four or eight Gaudi 2 accelerators.

Conclusion
In this blog post, we demonstrated the ease of Pristst inference and fine-tuning on the Gaudi 2, based on the optimality of Intel Gaudi Accelerators. Furthermore, our results show competitive performance against the A100, with a 1.76x speedup for inference and a 2.92x speedup for fine tuning. The following resources will help you get started with your Intel Gaudi 2 accelerator model:
Thank you for reading! With the capabilities of Intel Gaudi 2 Accelerator, we look forward to your innovation being built on the pioneer.



