the study
Published October 31, 2023 Author
Google DeepMind AlphaFold team and Isomorphic Labs team
Progress update: Our latest AlphaFold model has significantly improved accuracy and expanded coverage beyond proteins to other biomolecules, including ligands.
Since its release in 2020, AlphaFold has revolutionized the way we understand proteins and their interactions. Google DeepMind and Isomorphic Labs have collaborated to build the foundation for more powerful AI models that extend their reach beyond just proteins to the entire range of biologically relevant molecules.
Today we’re sharing an update on our progress towards the next generation of AlphaFold. Our state-of-the-art models can now generate predictions for almost any molecule in the Protein Data Bank (PDB), frequently reaching atomic precision.
This opens up new understanding of multiple major classes of biomolecules, including those involving ligands (small molecules), proteins, nucleic acids (DNA and RNA), and post-translational modifications (PTMs), and significantly increases precision. Masu. These different structural types and complexes are essential for understanding biological mechanisms within cells and have been difficult to predict with high accuracy.
The model’s enhanced functionality and performance will help accelerate biomedical breakthroughs and enable the next era of “digital biology.” This will enable new platforms to explore the functionality of disease pathways, genomics, biorenewable materials, plant immunity, potential therapeutic targets, drug discovery mechanisms, protein engineering and synthetic biology.
A set of predicted structures compared to the ground truth (white) of the latest AlphaFold model.
Beyond protein folding
AlphaFold was a fundamental advance in single-chain protein prediction. AlphaFold-Multimer was then extended to complexes with multiple protein chains, followed by AlphaFold2.3, which improved performance and extended applicability to larger complexes.
In 2022, AlphaFold structure predictions for nearly every cataloged protein known to science will be made freely available through the AlphaFold protein structure database in partnership with EMBL’s European Bioinformatics Institute (EMBL-EBI) Now you can.
To date, 1.4 million users in more than 190 countries have accessed the AlphaFold database, and scientists around the world are using AlphaFold’s predictions to discover everything from accelerating new malaria vaccines to advances in cancer drug discovery. We are conducting research in all fields, including the development of enzymes that eat plastic to combat pollution. .
Here we demonstrate AlphaFold’s remarkable ability to predict accurate structures beyond protein folding and generate highly accurate structural predictions across ligands, proteins, nucleic acids, and post-translational modifications.
Performance across protein-ligand complexes (a), proteins (b), nucleic acids (c), and covalent modifications (d).
Acceleration of drug discovery
Initial analyzes also show that our model significantly outperforms AlphaFold2.3 on several protein structure prediction problems relevant to drug discovery, such as antibody binding. Furthermore, accurately predicting protein-ligand structures is an extremely valuable tool for drug discovery, as it helps scientists identify and design new molecules that have the potential to become drugs.
The current industry standard is to use “docking methods” to determine interactions between ligands and proteins. These docking methods require a rigorous reference protein structure and recommended positions for the ligand to bind.
Our latest model sets a new standard for protein-ligand structure prediction by requiring no reference protein structure or ligand pocket location, and outperforming the best reported docking methods. Enables the prediction of completely novel proteins that have not been structurally characterized.
It also allows the positions of all atoms to be jointly modeled, allowing full expression of the inherent flexibility of proteins and nucleic acids as they interact with other molecules. This is not possible with docking techniques.
For example, here are three recently published cases related to treatment. In these cases, the structure predicted by the latest model (shown in color) is in good agreement with the experimentally determined structure (shown in gray).
PORCN: A clinical-stage anti-cancer molecule that binds to its target along with another protein. KRAS: a ternary complex with a covalent ligand (molecular glue) of an important cancer target. PI5P4Kγ: Selective allosteric inhibitor of lipid kinases. Impact on diseases such as cancer and immune diseases.
Prediction of PORCN (1), KRAS (2), and PI5P4Kγ (3).
Isomorphic Labs is applying this next-generation AlphaFold model to therapeutic drug design, contributing to the rapid and accurate characterization of many types of macromolecular structures important for disease treatment.
A new understanding of biology
By unlocking structural modeling of protein and ligand structures and nucleic acids, including post-translational modifications, our models provide faster and more accurate tools to interrogate fundamental biology.
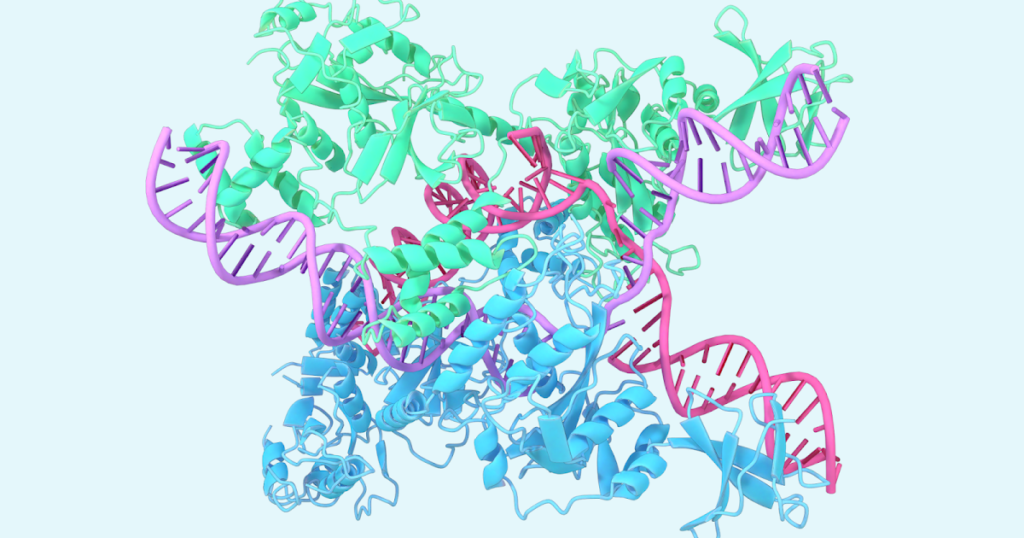
One example includes the structure of CasLambda bound to crRNA and DNA, which is part of the CRISPR family. CasLambda shares the genome editing power of the CRISPR-Cas9 system, commonly known as “genetic scissors,” which researchers can use to modify the DNA of animals, plants, and microorganisms. Due to CasLambda’s small size, it could potentially be used more efficiently in genome editing.
Prediction of the structure of CasLambda (Cas12l) bound to crRNA and DNA, which is part of the CRISPR subsystem.
The ability of the latest version of AlphaFold to model such complex systems shows that AI can help us better understand these types of mechanisms and accelerate their use for therapeutic applications. You can see more examples in our progress updates.
advances in scientific exploration
The dramatic jump in the performance of our model demonstrates the potential of AI to greatly enhance our scientific understanding of the molecular machines that make up the human body, and the broader natural world.
AlphaFold is already facilitating major scientific advances around the world. Now, the next generation of AlphaFold has the potential to help advance scientific exploration at digital speeds.
Our dedicated teams at Google DeepMind and Isomorphic Labs are making great progress on this important work and look forward to sharing our continued progress.